RESEARCH
Repeat Expansion Diseases
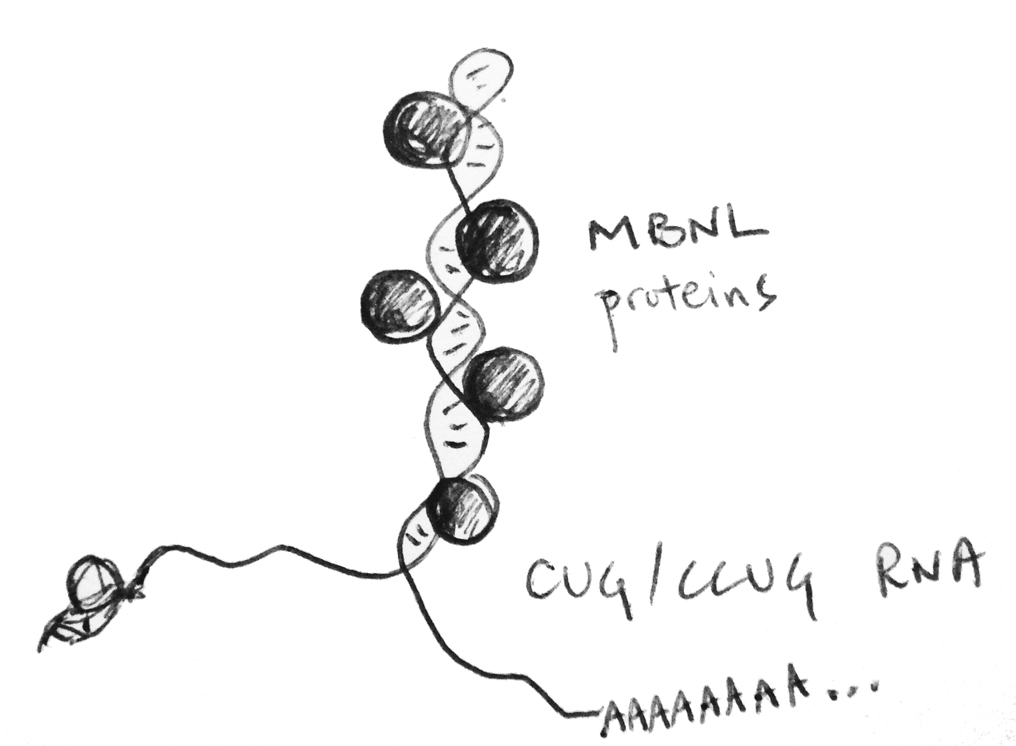 We are interested in all aspects of gene regulation and how its dysregulation causes
symptoms in microsatellite repeat expansion diseases. Myotonic dystrophy (dystrophia myotonica, DM) is one of these repeat expansion diseases and
has served as paradigm for a class of diseases caused by toxic RNA. We are using deep sequencing technologies, such as RNA-Seq and CLIP-Seq, in combination with molecular & cell biological techniques, to better understand how the repeating RNAs cause
a myriad of symptoms in virtually all tissues of the body, including skeletal, cardiac, and smooth muscle, and also tissues of the central nervous system.
We are interested in all aspects of gene regulation and how its dysregulation causes
symptoms in microsatellite repeat expansion diseases. Myotonic dystrophy (dystrophia myotonica, DM) is one of these repeat expansion diseases and
has served as paradigm for a class of diseases caused by toxic RNA. We are using deep sequencing technologies, such as RNA-Seq and CLIP-Seq, in combination with molecular & cell biological techniques, to better understand how the repeating RNAs cause
a myriad of symptoms in virtually all tissues of the body, including skeletal, cardiac, and smooth muscle, and also tissues of the central nervous system.
RNA Localization and Local Translation
 We found that the Muscleblind-like proteins (MBNLs), which are inactivated by the toxic RNA found in DM cells, also regulate the subcellular localization of many RNA species. We study how MBNL proteins can achieve these functions, and also how RNA localization is regulated at a basic level by RNA cis-elements and protein/RNA trans-factors. We aim to use both high-throughput, global approaches and high-resolution, single molecule approaches to define a "parts list" for RNA localization.
We found that the Muscleblind-like proteins (MBNLs), which are inactivated by the toxic RNA found in DM cells, also regulate the subcellular localization of many RNA species. We study how MBNL proteins can achieve these functions, and also how RNA localization is regulated at a basic level by RNA cis-elements and protein/RNA trans-factors. We aim to use both high-throughput, global approaches and high-resolution, single molecule approaches to define a "parts list" for RNA localization.
Development of Therapeutic Approaches
Our studies of DM pathogenesis and RNA regulation are in part driven by a desire to find treatments for DM and other related diseases. We work with diverse partners in both the academic and industrial space to make progress in this area, and are taking a number of approaches to develop therapeutics that may one day enter the clinic.
Publications
2018
- Eric T Wang, Daniel Treacy, Katy Eichinger, Adam Struck, Joseph Estabrook, Hailey Olafson, Thomas T Wang, Kirti Bhatt, Tony Westbrook, Sam Sedehizadeh, Amanda Ward, John Day, David Brook, J Andrew Berglund, Thomas Cooper, David Housman, Charles Thornton and Christopher Burge. Transcriptome alterations in myotonic dystrophy skeletal muscle and heart. Hum Mol Genet, 2018.
- Fatma Ayhan, Barbara A Perez, Hannah K Shorrock, Tao Zu, Monica Banez-Coronel, Tammy Reid, Hirokazu Furuya, H Brent Clark, Juan C Troncoso, Christopher A Ross, S H Subramony, Tetsuo Ashizawa, Eric T Wang, Anthony T Yachnis and Laura Pw Ranum. SCA8 RAN polySer protein preferentially accumulates in white matter regions and is regulated by eIF3F. EMBO J, 37 (19), 2018.
- Pietro Fratta, Prasanth Sivakumar, Jack Humphrey, Kitty Lo, Thomas Ricketts, Hugo Oliveira, Jose M Brito-Armas, Bernadett Kalmar, Agnieszka Ule, Yichao Yu, Nicol Birsa, Cristian Bodo, Toby Collins, Alexander E Conicella, Alan Mejia Maza, Alessandro Marrero-Gagliardi, Michelle Stewart, Joffrey Mianne, Silvia Corrochano, Warren Emmett, Gemma Codner, Michael Groves, Ryutaro Fukumura, Yoichi Gondo, Mark Lythgoe, Erwin Pauws, Emma Peskett, Philip Stanier, Lydia Teboul, Martina Hallegger, Andrea Calvo, Adriano Chi`o, Adrian M Isaacs, Nicolas L Fawzi, Eric Wang, David E Housman, Francisco Baralle, Linda Greensmith, Emanuele Buratti, Vincent Plagnol, Elizabeth Mc Fisher and Abraham Acevedo-Arozena. Mice with endogenous TDP-43 mutations exhibit gain of splicing function and characteristics of amyotrophic lateral sclerosis. EMBO J, 37 (11), 2018.
- Melissa A Hale, Jared I Richardson, Ryan C Day, Ona L McConnell, Juan Arboleda, Eric T Wang and J Andrew Berglund. An engineered RNA binding protein with improved splicing regulation. Nucleic Acids Res, 46 (6): 3152-3168, 2018.
2017
- Masayuki Nakamori, Kohei Hamanaka, James D Thomas, Eric T Wang, Yukiko K Hayashi, Masanori P Takahashi, Maurice S Swanson, Ichizo Nishino and Hideki Mochizuki. Aberrant Myokine Signaling in Congenital Myotonic Dystrophy. Cell Rep, 21 (5): 1240-1252, 2017.
- Belinda S Pinto, Tanvi Saxena, Ruan Oliveira, H'ector R M'endez-G'omez, John D Cleary, Lance T Denes, Ona McConnell, Juan Arboleda, Guangbin Xia, Maurice S Swanson and Eric T Wang. Impeding Transcription of Expanded Microsatellite Repeats by Deactivated Cas9. Mol Cell, 68 (3): 479-490.e5, 2017.
- Anke E E G Gudde, Simon J van Heeringen, Amanda I de Oude, Ingeborg D G van Kessel, Joseph Estabrook, Eric T Wang, B'e Wieringa and Derick G Wansink. Antisense transcription of the myotonic dystrophy locus yields low-abundant RNAs with and without (CAG)n repeat. RNA Biol, 14 (10): 1374-1388, 2017.
- Ryan J McGinty, Franco Puleo, Anna Y Aksenova, Julia A Hisey, Alexander A Shishkin, Erika L Pearson, Eric T Wang, David E Housman, Claire Moore and Sergei M Mirkin. A Defective mRNA Cleavage and Polyadenylation Complex Facilitates Expansions of Transcribed (GAA)n Repeats Associated with Friedreich's Ataxia. Cell Rep, 20 (10): 2490-2500, 2017.
- Fan Zhang, Nicole E Bodycombe, Keith M Haskell, Yumei L Sun, Eric T Wang, Carl A Morris, Lyn H Jones, Lauren D Wood and Mathew T Pletcher. A flow cytometry-based screen identifies MBNL1 modulators that rescue splicing defects in myotonic dystrophy type I. Hum Mol Genet, 26 (16): 3056-3068, 2017.
- James D Thomas, Lukasz J Sznajder, Olgert Bardhi, Faaiq N Aslam, Zacharias P Anastasiadis, Marina M Scotti, Ichizo Nishino, Masayuki Nakamori, Eric T Wang and Maurice S Swanson. Disrupted prenatal RNA processing and myogenesis in congenital myotonic dystrophy. Genes Dev, 31 (11): 1122-1133, 2017.
- Charles A Thornton, Eric Wang and Ellie M Carrell. Myotonic dystrophy: approach to therapy. Curr Opin Genet Dev, 44: 135-140, 2017.
- Kevin Yum, Eric T Wang and Auinash Kalsotra. Myotonic dystrophy: disease repeat range, penetrance, age of onset, and relationship between repeat size and phenotypes. Curr Opin Genet Dev, 44: 30-37, 2017.
- James E Dahlman, Kevin J Kauffman, Yiping Xing, Taylor E Shaw, Faryal F Mir, Chloe C Dlott, Robert Langer, Daniel G Anderson and Eric T Wang. Barcoded nanoparticles for high throughput in vivo discovery of targeted therapeutics. Proc Natl Acad Sci U S A, 114 (8): 2060-2065, 2017.
- Marina Vidaki, Frauke Drees, Tanvi Saxena, Erwin Lanslots, Matthew J Taliaferro, Antonios Tatarakis, Christopher B Burge, Eric T Wang and Frank B Gertler. A Requirement for Mena, an Actin Regulator, in Local mRNA Translation in Developing Neurons. Neuron, 95 (3): 608-622.e5, 2017.
2016
- Genevieve M Gould, Joseph M Paggi, Yuchun Guo, David V Phizicky, Boris Zinshteyn, Eric T Wang, Wendy V Gilbert, David K Gifford and Christopher B Burge. Identification of new branch points and unconventional introns in Saccharomyces cerevisiae. RNA, 22 (10): 1522-34, 2016.
- Stacey D Wagner, Adam J Struck, Riti Gupta, Dylan R Farnsworth, Amy E Mahady, Katy Eichinger, Charles A Thornton, Eric T Wang and J Andrew Berglund. Dose-Dependent Regulation of Alternative Splicing by MBNL Proteins Reveals Biomarkers for Myotonic Dystrophy. PLoS Genet, 12 (9): e1006316, 2016.
- Julia C Oddo, Tanvi Saxena, Ona L McConnell, J Andrew Berglund and Eric T Wang. Conservation of context-dependent splicing activity in distant Muscleblind homologs. Nucleic Acids Res, 44 (17): 8352-62, 2016.
- Fernande Freyermuth, Fr'ed'erique Rau, Yosuke Kokunai, Thomas Linke, Chantal Sellier, Masayuki Nakamori, Yoshihiro Kino, Ludovic Arandel, Arnaud Jollet, Christelle Thibault, Muriel Philipps, Serge Vicaire, Bernard Jost, Bjarne Udd, John W Day, Denis Duboc, Karim Wahbi, Tsuyoshi Matsumura, Harutoshi Fujimura, Hideki Mochizuki, Franc cois Deryckere, Takashi Kimura, Nobuyuki Nukina, Shoichi Ishiura, Vincent Lacroix, Amandine Campan-Fournier, Vincent Navratil, Emilie Chautard, Didier Auboeuf, Minoru Horie, Keiji Imoto, Kuang-Yung Lee, Maurice S Swanson, Adolfo Lopez de Munain, Shin Inada, Hideki Itoh, Kazuo Nakazawa, Takashi Ashihara, Eric Wang, Thomas Zimmer, Denis Furling, Masanori P Takahashi and Nicolas Charlet-Berguerand. Splicing misregulation of SCN5A contributes to cardiac-conduction delay and heart arrhythmia in myotonic dystrophy. Nat Commun, 7: 11067, 2016.
- J Matthew Taliaferro, Marina Vidaki, Ruan Oliveira, Sara Olson, Lijun Zhan, Tanvi Saxena, Eric T Wang, Brenton R Graveley, Frank B Gertler, Maurice S Swanson and Christopher B Burge. Distal Alternative Last Exons Localize mRNAs to Neural Projections. Mol Cell, 61 (6): 821-33, 2016.
- Eric T Wang, J Matthew Taliaferro, Ji-Ann Lee, Indulekha P Sudhakaran, Wilfried Rossoll, Christina Gross, Kathryn R Moss and Gary J Bassell. Dysregulation of mRNA Localization and Translation in Genetic Disease. J Neurosci, 36 (45): 11418-11426, 2016.
2015
- Eric T Wang, Amanda J Ward, Jennifer Cherone, Thomas T Wang, Jimena Giudice, Daniel Treacy, Peter Freese, Nicole J Lambert, Tanvi Saxena, Thomas A Cooper and Christopher B Burge. Antagonistic Regulation of mRNA Expression and Splicing by CELF and MBNL Proteins. Genome Res, 2015.
- Yarden Katz, Eric T Wang, Jacob Silterra, Schraga Schwartz, Bang Wong, Helga Thorvaldsd'ottir, James T Robinson, Jill P Mesirov, Edoardo M Airoldi and Christopher B Burge. Quantitative visualization of alternative exon expression from RNA-seq data. Bioinformatics, 2015.
2014
- Jimena Giudice, Zheng Xia, Eric T Wang, Marissa A Scavuzzo, Amanda J Ward, Auinash Kalsotra, Wei Wang, Xander H T Wehrens, Christopher B Burge, Wei Li and Thomas A Cooper. Alternative splicing regulates vesicular trafficking genes in cardiomyocytes during postnatal heart development. Nat Commun, 5: 3603, 2014.
- Albert W Cheng, Jiahai Shi, Piu Wong, Katherine L Luo, Paula Trepman, Eric T Wang, Heejo Choi, Christopher B Burge and Harvey F Lodish. Muscleblind-like 1 (Mbnl1) regulates pre-mRNA alternative splicing during terminal erythropoiesis. Blood, 124 (4): 598-610, 2014.
- J Matthew Taliaferro, Eric T Wang and Christopher B Burge. Genomic analysis of RNA localization. RNA Biol, 11 (8): 1040-50, 2014.
2013
- Hong Han, Manuel Irimia, P Joel Ross, Hoon-Ki Sung, Babak Alipanahi, Laurent David, Azadeh Golipour, Mathieu Gabut, Iacovos P Michael, Emil N Nachman, Eric Wang, Dan Trcka, Tadeo Thompson, Dave O'Hanlon, Valentina Slobodeniuc, Nuno L Barbosa-Morais, Christopher B Burge, Jason Moffat, Brendan J Frey, Andras Nagy, James Ellis, Jeffrey L Wrana and Benjamin J Blencowe. MBNL proteins repress ES-cell-specific alternative splicing and reprogramming. Nature, 498 (7453): 241-5, 2013.
- Marka van Blitterswijk, Eric T Wang, Brad A Friedman, Pamela J Keagle, Patrick Lowe, Ashley Lyn Leclerc, Leonard H van den Berg, David E Housman, Jan H Veldink and John E Landers. Characterization of FUS mutations in amyotrophic lateral sclerosis using RNA-Seq. PLoS One, 8 (4): e60788, 2013.
- Mohammadsharif Tabebordbar, Eric T Wang and Amy J Wagers. Skeletal muscle degenerative diseases and strategies for therapeutic muscle repair. Annu Rev Pathol, 8: 441-75, 2013.
2012
- Raman Parkesh, Jessica L Childs-Disney, Masayuki Nakamori, Amit Kumar, Eric Wang, Thomas Wang, Jason Hoskins, Tuan Tran, David Housman, Charles A Thornton and Matthew D Disney. Design of a bioactive small molecule that targets the myotonic dystrophy type 1 RNA via an RNA motif-ligand database and chemical similarity searching. J Am Chem Soc, 134 (10): 4731-42, 2012.
- Jamie Purcell, Julia C Oddo, Eric T Wang and J Andrew Berglund. Combinatorial Mutagenesis of MBNL1 Zinc Fingers Elucidates Distinct Classes of Regulatory Events. Mol Cell Biol, 32 (20): 4155-67, 2012.
- Maja M Janas, Eric Wang, Tara Love, Abigail S Harris, Kristen Stevenson, Karlheinz Semmelmann, Jonathan M Shaffer, Po-Hao Chen, John G Doench, Subrahmanyam V B K Yerramilli, Donna S Neuberg, Dimitrios Iliopoulos, David E Housman, Christopher B Burge and Carl D Novina. Reduced expression of ribosomal proteins relieves microRNA-mediated repression. Mol Cell, 46 (2): 171-86, 2012.
- Eric T Wang, Neal A L Cody, Sonali Jog, Michela Biancolella, Thomas T Wang, Daniel J Treacy, Shujun Luo, Gary P Schroth, David E Housman, Sita Reddy, Eric L'ecuyer and Christopher B Burge. Transcriptome-wide Regulation of Pre-mRNA Splicing and mRNA Localization by Muscleblind Proteins. Cell, 150 (4): 710-24, 2012.
2010
- Christopher S Bland, Eric T Wang, Anthony Vu, Marjorie P David, John C Castle, Jason M Johnson, Christopher B Burge and Thomas A Cooper. Global regulation of alternative splicing during myogenic differentiation. Nucleic Acids Res, 38 (21): 7651-64, 2010.
- Yarden Katz, Eric T Wang, Edoardo M Airoldi and Christopher B Burge. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods, 7 (12): 1009-15, 2010.
2009
- Xinshu Xiao, Zefeng Wang, Minyoung Jang, Razvan Nutiu, Eric T Wang and Christopher B Burge. Splice site strength-dependent activity and genetic buffering by poly-G runs. Nat Struct Mol Biol, 16 (10): 1094-100, 2009.
- Daniel Ramsk"old, Eric T Wang, Christopher B Burge and Rickard Sandberg. An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Comput Biol, 5 (12): e1000598, 2009.
2008
- Eric T Wang, Rickard Sandberg, Shujun Luo, Irina Khrebtukova, Lu Zhang, Christine Mayr, Stephen F Kingsmore, Gary P Schroth and Christopher B Burge. Alternative isoform regulation in human tissue transcriptomes. Nature, 456 (7221): 470-6, 2008.
2007
- Guohao Dai, Saran Vaughn, Yuzhi Zhang, Eric T Wang, Guillermo Garcia-Cardena and Jr, Michael A Gimbrone. Biomechanical forces in atherosclerosis-resistant vascular regions regulate endothelial redox balance via phosphoinositol 3-kinase/Akt-dependent activation of Nrf2. Circ Res, 101 (7): 723-33, 2007.
2006
- Kush M Parmar, H Benjamin Larman, Guohao Dai, Yuzhi Zhang, Eric T Wang, Sripriya N Moorthy, Johannes R Kratz, Zhiyong Lin, Mukesh K Jain, Jr, Michael A Gimbrone and Guillermo Garcia-Cardena. Integration of flow-dependent endothelial phenotypes by Kruppel-like factor 2. J Clin Invest, 116 (1): 49-58, 2006.
- Deborah Abeles, Stephanie Kwei, George Stavrakis, Yuzhi Zhang, Eric T Wang and Guillermo Garcia-Cardena. Gene expression changes evoked in a venous segment exposed to arterial flow. J Vasc Surg, 44 (4): 863-70, 2006.
 We are interested in all aspects of gene regulation and how its dysregulation causes
symptoms in microsatellite repeat expansion diseases. Myotonic dystrophy (dystrophia myotonica, DM) is one of these repeat expansion diseases and
has served as paradigm for a class of diseases caused by toxic RNA. We are using deep sequencing technologies, such as RNA-Seq and CLIP-Seq, in combination with molecular & cell biological techniques, to better understand how the repeating RNAs cause
a myriad of symptoms in virtually all tissues of the body, including skeletal, cardiac, and smooth muscle, and also tissues of the central nervous system.
We are interested in all aspects of gene regulation and how its dysregulation causes
symptoms in microsatellite repeat expansion diseases. Myotonic dystrophy (dystrophia myotonica, DM) is one of these repeat expansion diseases and
has served as paradigm for a class of diseases caused by toxic RNA. We are using deep sequencing technologies, such as RNA-Seq and CLIP-Seq, in combination with molecular & cell biological techniques, to better understand how the repeating RNAs cause
a myriad of symptoms in virtually all tissues of the body, including skeletal, cardiac, and smooth muscle, and also tissues of the central nervous system.
 We found that the Muscleblind-like proteins (MBNLs), which are inactivated by the toxic RNA found in DM cells, also regulate the subcellular localization of many RNA species. We study how MBNL proteins can achieve these functions, and also how RNA localization is regulated at a basic level by RNA cis-elements and protein/RNA trans-factors. We aim to use both high-throughput, global approaches and high-resolution, single molecule approaches to define a "parts list" for RNA localization.
We found that the Muscleblind-like proteins (MBNLs), which are inactivated by the toxic RNA found in DM cells, also regulate the subcellular localization of many RNA species. We study how MBNL proteins can achieve these functions, and also how RNA localization is regulated at a basic level by RNA cis-elements and protein/RNA trans-factors. We aim to use both high-throughput, global approaches and high-resolution, single molecule approaches to define a "parts list" for RNA localization.